Artificial intelligence (AI) and machine learning (ML) are beginning to reshape healthcare delivery and discovery. CHIP researchers work across the spectrum from predictive medicine, to digital disease detection, to image processing.
Working Groups
CHIP, in partnerships with other programs, has led the launch of Boston Children’s Hospital Artificial Intelligence and Machine Learning Working Group, which gives Boston Children’s clinicians and investigators a forum for sharing knowledge and collaborating across the many facets of artificial intelligence and machine learning.
Projects

CHIP has designed and implemented Digital Disease Detection tools as decision-support tools to help public health officials make timely decisions and prevent epidemic outbreaks.

CHIP conducts research to understand the impact of climate change on disease burden in Africa and Mexico.

The availability of online and real-time sources of big data provides the opportunity to monitor how climate variables influence the spread of Dengue, Antibiotic Resistance, and other outbreaks around the world.

Mathematical concepts of machine learning are applied to improve patient outcomes and reduce hospital costs in Critical Care Medicine.

Understanding the global-scale dynamics of the chemical composition of our atmosphere is essential for addressing a wide range of environmental issues from air quality to climate change. Understanding this phenomenon enables us to evaluate and devise appropriate environmental policies, such as the Kyoto Protocol on global greenhouse gases emissions.

As a consequence of our changing climate, large efforts have been made to understand the social risks of storm surges (hypothesized to increase in frequency in warmer climate scenarios) and sea level rise in coastal areas. Of particular interest is the role that wetlands and coastal marshes play in storm surges and flooding events.

The Intelligent Histories project develops new ways of using commonly available electronic medical information to predict people's future medical risks, helping doctors choose preventive interventions and improve medical care.

Suicide is one of the ten leading causes of death in the United States. Even though the majority of all individuals who die by suicide have contact with a healthcare professional in the month before their death, suicide risk is rarely detected in such cases. CHIP researchers have developed advanced predictive models able to identify between one third and one half of all suicide attempts on average three years before they occur, enabling life-saving interventions and care.

Psychosis often first appears during adolescence or young adulthood and is difficult to detect. If left undetected and untreated, psychosis can quickly deteriorate to even more severe mental illness. CHIP researchers are developing advanced predictive models to identify cases of first episode psychosis years prior to when they would otherwise be detected by the health system.

Family histories are an essential predictor of disease risk, yet they are often incomplete, inaccurate, and underutilized in today's clinical settings. CHIP researchers are developing improved approaches to providing more complete, accurate and detailed family histories based on electronic health records of patients and their consenting family members. These improved histories enable better clinical risk prediction and decision-making.

CHIP researchers have developed novel network-based models to predict unknown adverse drug events and drug-drug interactions. Instead of waiting for sufficient post-marketing evidence to accumulate, this predictive approach can identify drug safety issues years in advance.

CHIP researchers have developed new ways of using commonly available electronic medical information to predict people's future medical risks helping doctors choose preventive interventions and improve medical care.
We apply advanced modeling techniques to novel data sources in order to predict and detect outbreaks and other public health trends, especially during times of great uncertainty such as epidemics or large public events
All of our work with the Epilepsy Center has a clear clinical focus, mostly around discovery of biomarkers for epilepsy, comorbid neurological or cognitive effects, and seizure detection. Our primary focus is on developing and implementing new methods for nonlinear EEG signal analysis, data integration, data structures for complex feature extraction, and machine learning for classification of epilepsy or seizures.

We have demonstrated that biomarkers for emerging Autism Spectrum Disorder can be computed from EEG signals as early as 3 months of age. Our work continues with further testing in a general population of children in routine pediatric checkups.
We developed a crowdsourcing platform for EEG annotation and accuracy estimation. In related research that involves software and data engineering, we are beginning work on an EEG Biobank that will make BCH clinical EEG data available for researchers.

Our research in neuroinformatics is based on a nonlinear systems approach to analyzing EEG signals, and integration of the derived signal information with other physiological or clinical data. Research involves nonlinear dynamical systems theory, recurrence plot analysis, synchronization methods, tensor formulation of complex biomarker data, and machine learning for classification.

The Prediction of Patient Placement (POPP) system aims to improve the flow of patients through the Emergency Department (ED) and hospital, by providing decision makers with real-time predictions of future patient disposition. POPP bridges predictive analytics to the point of care. We apply computer models on live data extracted from the Electronic Health Records to forecast not only what patients currently need, but what will they need in the near future - facilitating a smarter and more efficient use of resources. Building upon our predictive models we developed a Dashboard.

Apache clinical Text Analysis and Knowledge Extraction System (cTAKES) is a widely used, open source and free tool for clinical natural language processing (NLP). Unlike general purpose NLP tools, cTAKES is specialized for clinical texts, incorporating Unified Medical Language System (UMLS) resources for finding medical concepts and packaged with machine learning models trained on gold standard clinical texts. Apache cTAKES has NLP use that extends beyond clinical care. Apache cTAKES became the first and only top-level Apache Software Foundation biomedical informatics software in 2013. In 2019, Apache cTAKES was named one of the 20 most influential Apache projects.

CHIP researchers develop novel methods for information extraction to facilitate automatic/unsupervised/minimally supervised extraction of specific discrete cancer- related data from various types of unstructured electronic medical records. Our two main use cases are cancer deep phenotyping for translational science (DeepPhe) and a platform for cancer surveillance by the cancer registries (DeepPhe*CR).

Temporal Histories of Your Medical Event (THYME) uses temporal relations in processing free text. Understanding the timeline of clinically relevant events is key to the next generation of translational research where the importance of generalizing over large amounts of data holds the promise of deciphering biomedical puzzles.

The Health Natural Language Processing (hNLP) Center targets a key challenge to current hNLP research and health-related human language technology development: the lack of health-related language data. The Center’s primary activities are to: provide a repository and a data curation, distribution and management point for health-related language resources, support sponsored research programs and health-related language-based technology evaluations, and engage in collaborations with US and foreign researchers, institutions and data centers.

Our goals are to apply the best performing NLP methods to impactful biomedical uses cases to advance the science of biomedicine and clinical care, such as, pediatric pulmonary hypertension, rheumatoid arthritis, inflammatory bowel disease, artery aneurysms, early childhood obesity, autism spectrum disorder, polycystic ovary syndrome, and methotrexate-induced liver toxicity.
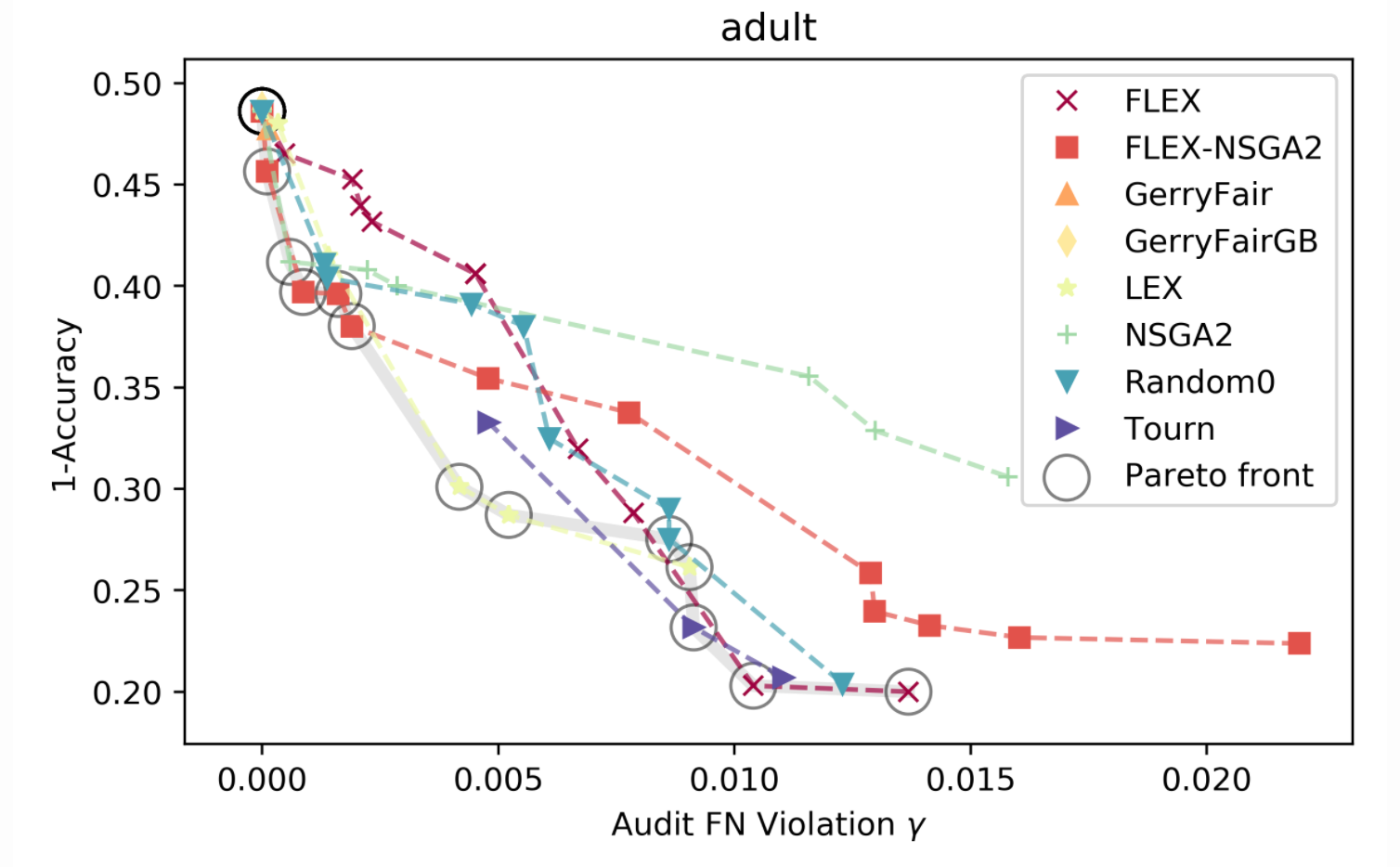
The Fairness in Machine Learning project focuses on addressing these challenges by developing flexible search methods that can explicitly optimize multiple criteria - in this case, between model accuracy and fairness. Our faculty are interested in understanding the intricacies of downstream impacts on healthcare that will arise as more and more models are deployed in the health system. Providing a set of models varying in fairness and accuracy is one way to aid a decision maker in understanding how an algorithm will affect the people it interacts with when it is deployed.
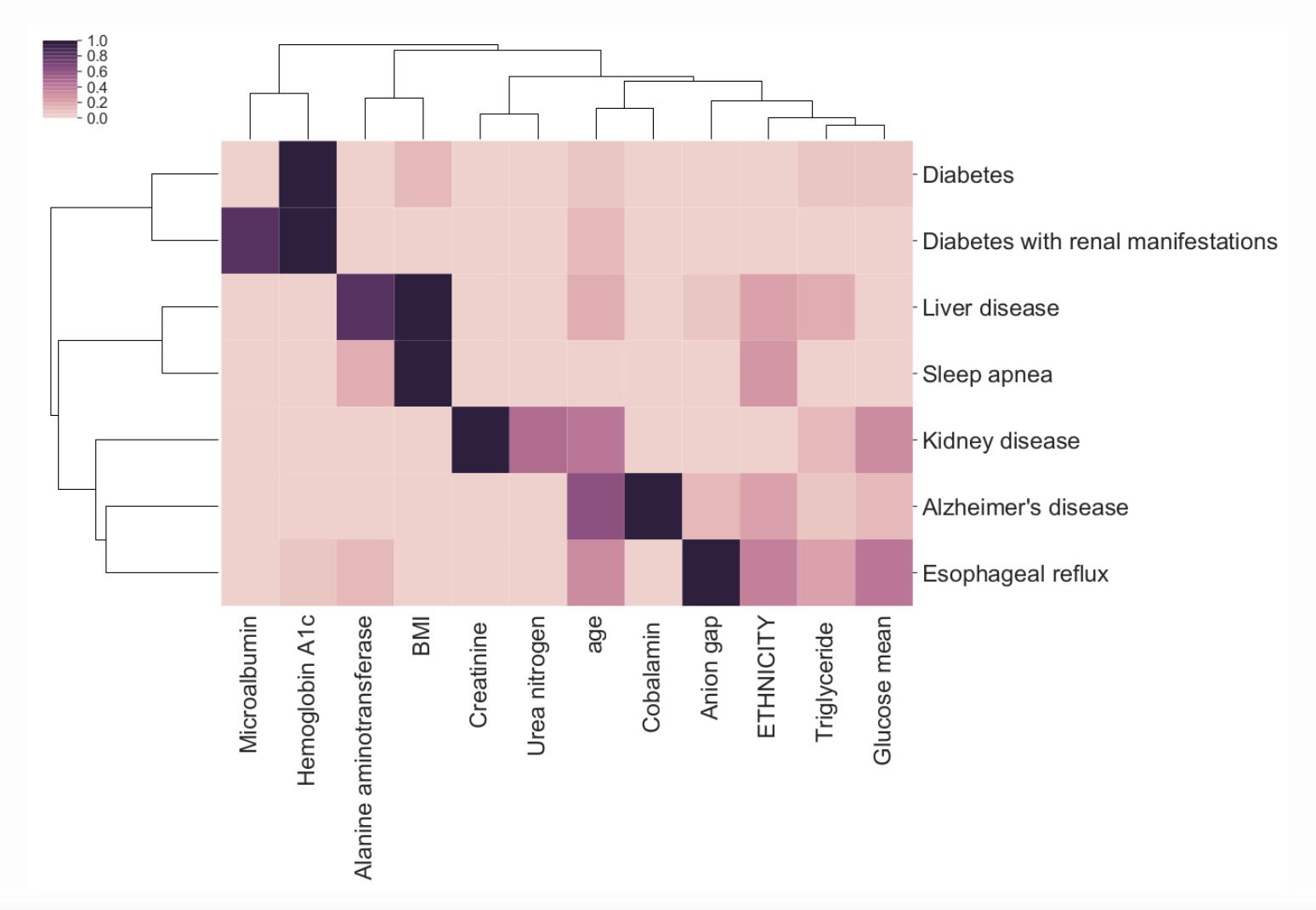
Intelligible Predictive Health Models seeks to analyze and improve the intelligibility and/or explainability of Machine Learning (ML) methods deployed in the health system. We empirically analyze how intelligible current ML tools are and develop novel approaches to interpretable ML.
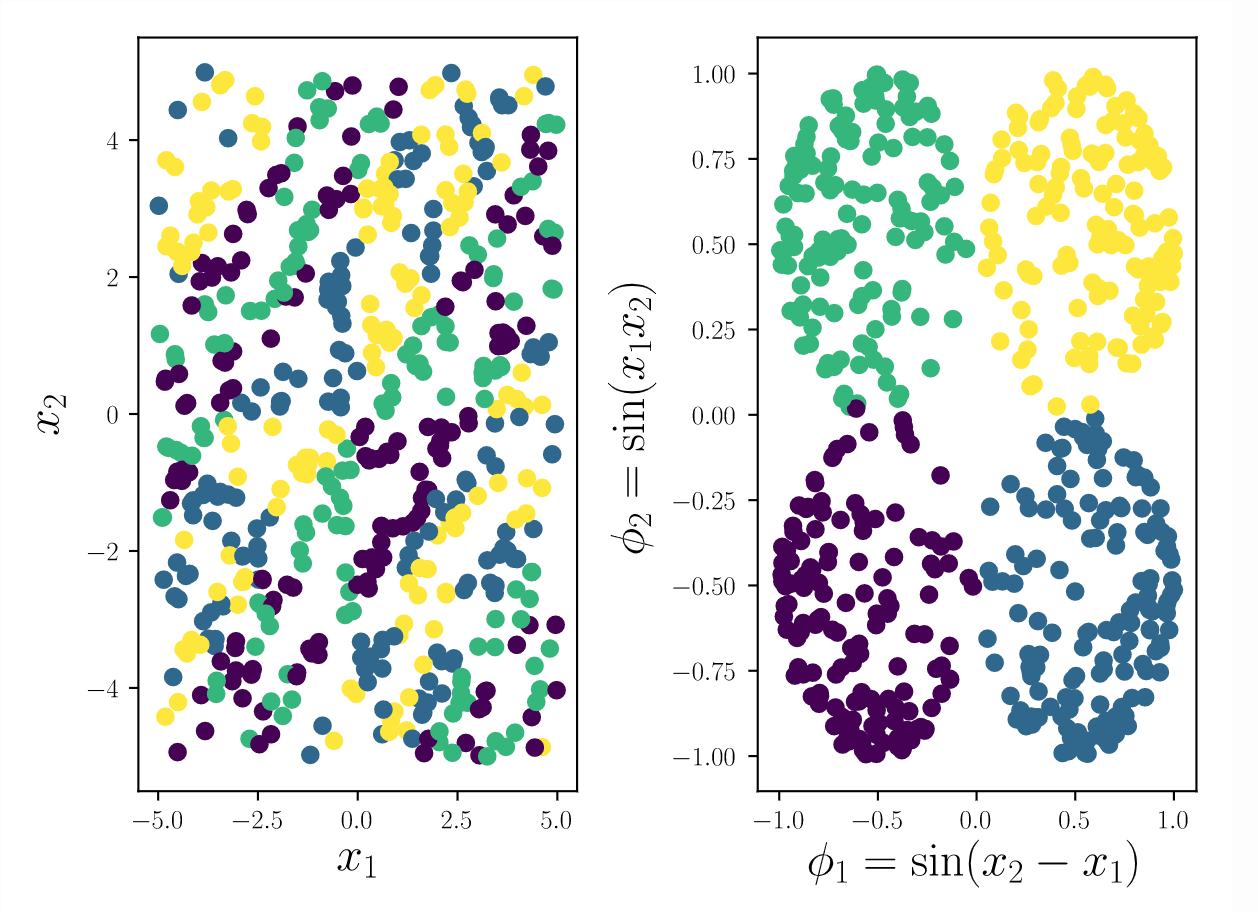
What makes a representation, i.e. a feature space, good? At the minimum, a good representation produces a model with better generalization than a model trained only on the raw data attributes. In addition, a good representation teases apart the factors of variation in the data into independent components. Finally, an ideal representation is succinct so as to promote intelligibility. This means a representation should only have as many features as there are independent factors in the process, and each of those features should be digestible by the user. This research project centers around these three motivations when designing novel algorithms for interpretable machine learning.